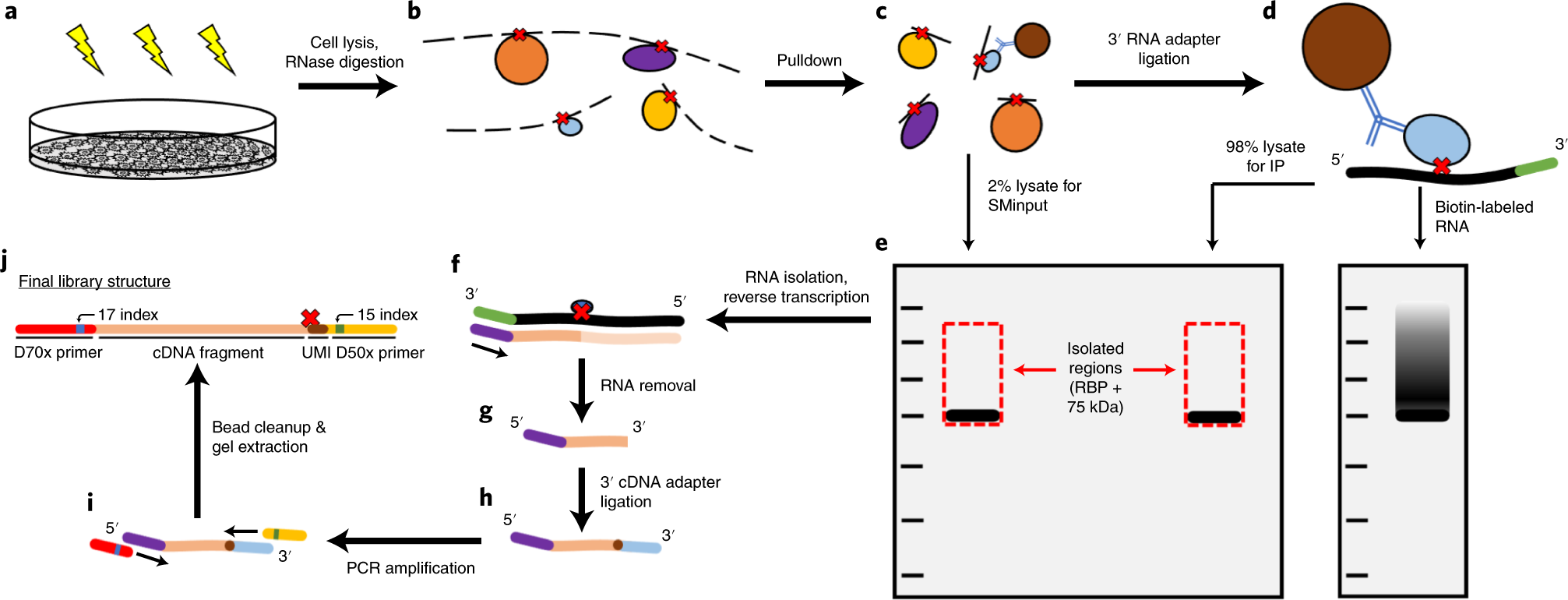
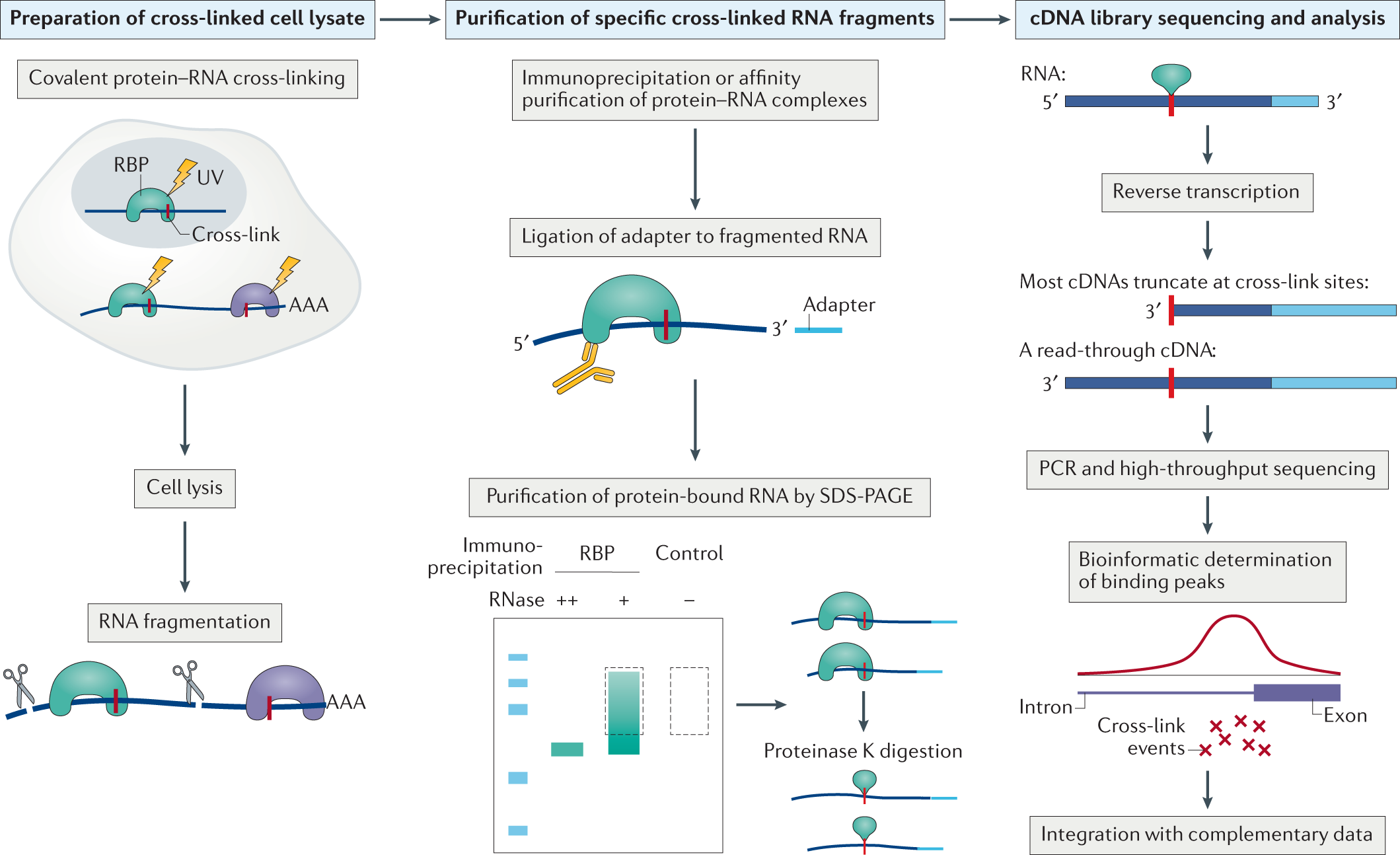
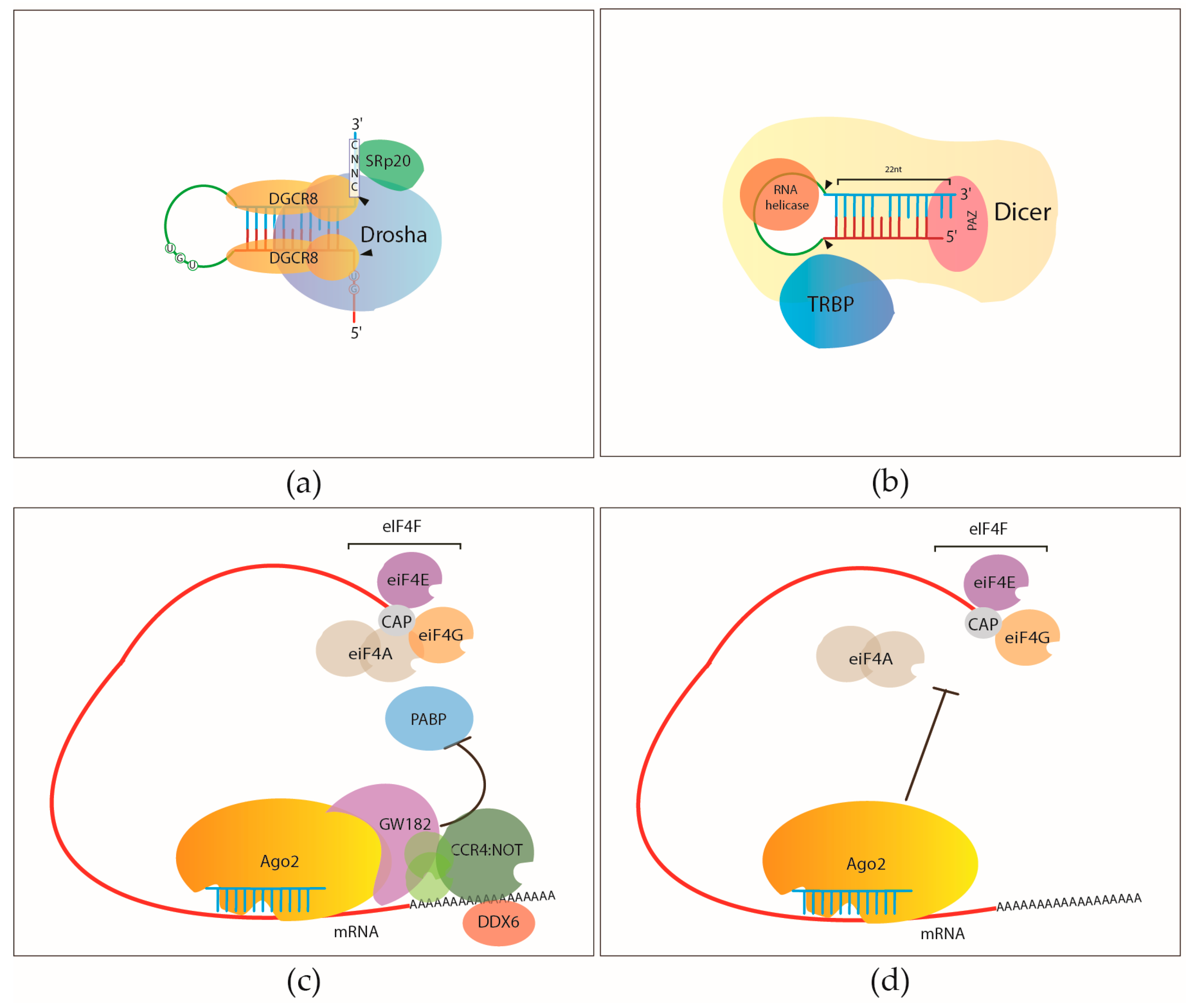
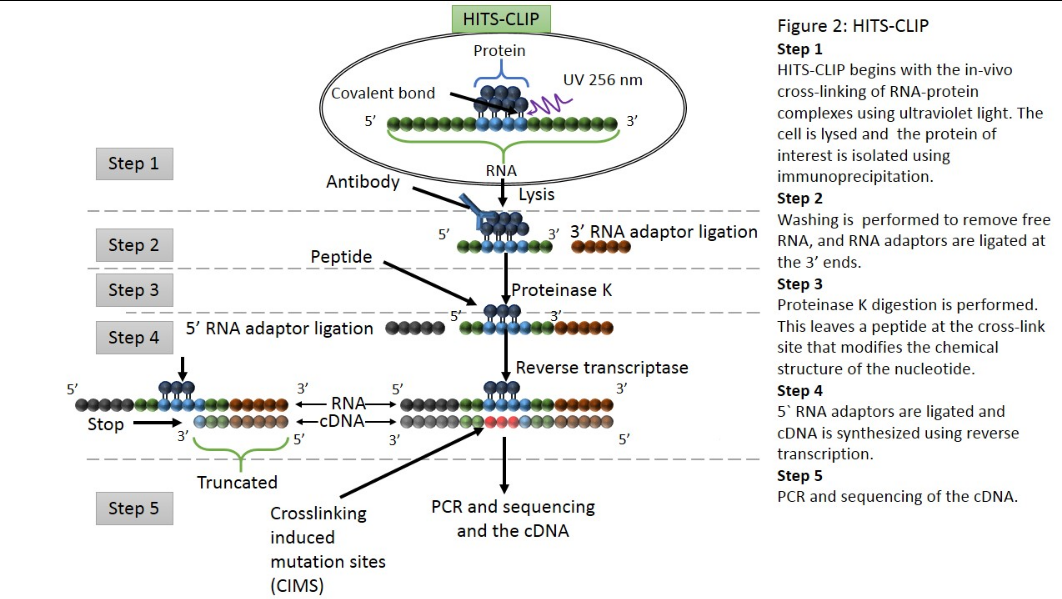
Argonaute-CLIP delineates versatile, functional RNAi networks in Aedes aegypti, a major vector of human viruses - ScienceDirect

CLIP-seq overview. The CLIP-seq method for identifying miRNA target... | Download Scientific Diagram
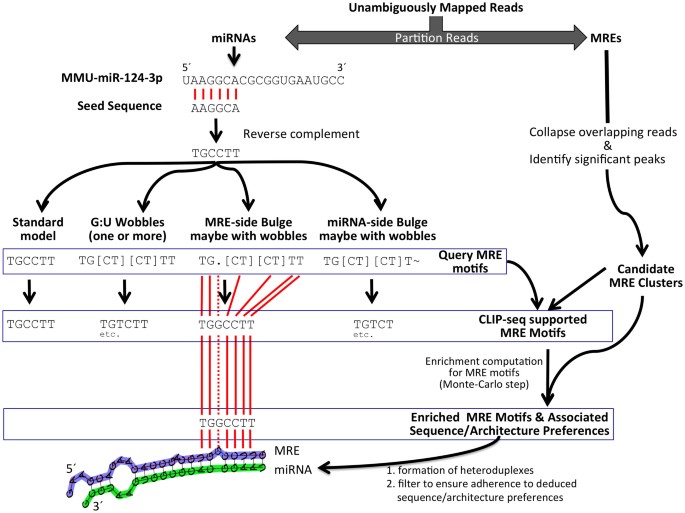
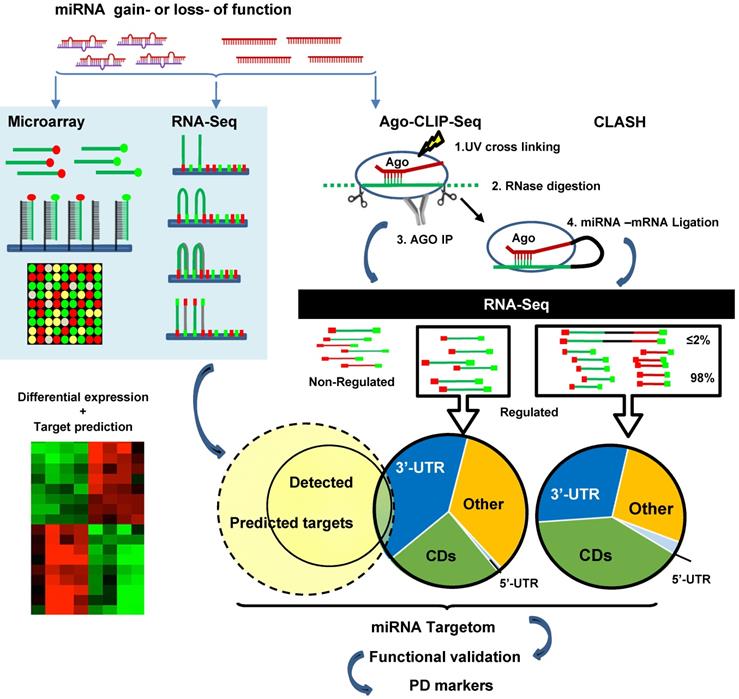
Learning to Predict miRNA-mRNA Interactions from AGO CLIP Sequencing and CLASH Data | PLOS Computational Biology
starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data

circTools: identify and annotate circRNAs and their interactions with microRNAs (miRNAs) from large-scale <strong>CLIP-Seq (HITS-CLIP, PAR-CLIP, iCLIP, CLASH)</strong> and RNA-seq data.
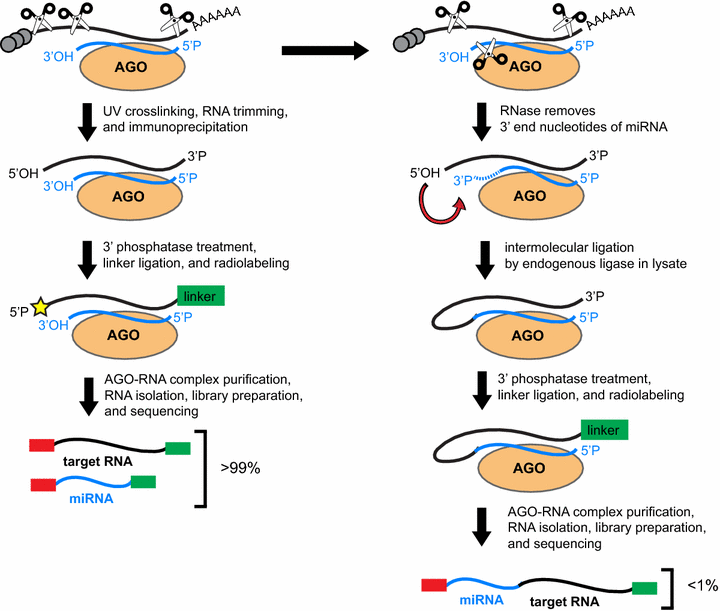
Sequencing of Argonaute-bound microRNA/mRNA hybrids reveals regulation of the unfolded protein response by microRNA-320a | PLOS Genetics

AGO CLIP Reveals an Activated Network for Acute Regulation of Brain Glutamate Homeostasis in Ischemic Stroke - ScienceDirect

Interpretation of the AGO-binding model learned from CLIP-seq data. (A)... | Download Scientific Diagram

Discovering the Interactions between Circular RNAs and RNA-binding Proteins from CLIP-seq Data using circScan | bioRxiv

![PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6bbd241680ec0f3d6a86dec738a6b864ea5048c3/2-Figure1-1.png)